
Under the normality assumption, Section 1.05.4 summarizes the confidence intervals and the inferential procedures needed for an adequate treatment of the analytical questions, such as the uncertainty of the analytical determinations, the comparison of calibration models, or the evaluation of the bias. The assessment of the hypotheses of the model is the subject of Section 1.05.3. Section 1.05.1 gives a brief introduction, and Section 1.05.2 introduces the notation and the univariate linear regression model related to the calibration, and shows the least squares solution and the effect of supposing a normal distribution for the errors. This is the basic objective of this chapter.
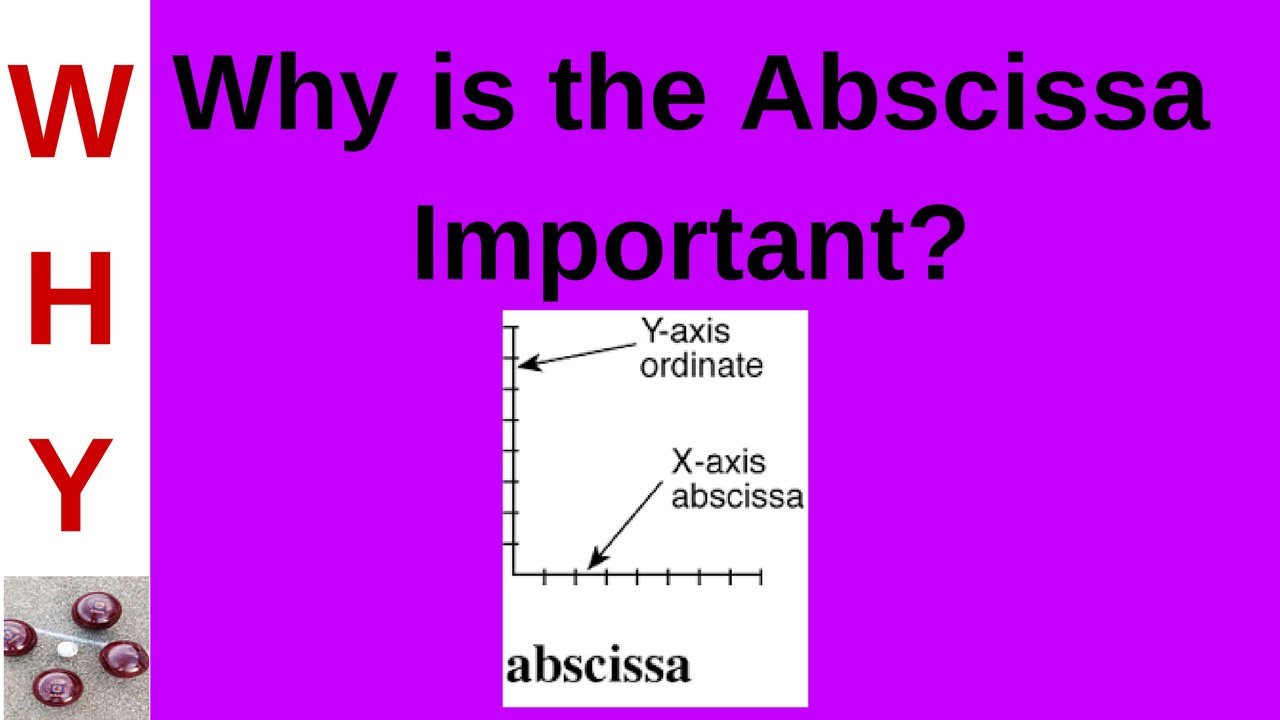
ABSCISSA VALUES HOW TO
After that, this relationship is used to transform the measurements made in a test sample into estimates of the quantity of the analyte.Ĭonsequently, it is necessary that the analysts have a good understanding of how to carry out a calibration experiment and how to evaluate the results obtained. In general, the calibration of an instrument supposes to prepare a set of calibration standards with known quantity of the analyte of interest, to measure the response of the instrument for each standard, and to establish the relationship between the response of the instrument and the concentration of the analyte. VF estimation is illustrated with an example from the recent literature.įrom a statistical point of view, the regression analysis is an area of ongoing research, so an ever-growing collection of techniques makes it difficult the selection of the most adequate one for a given problem.įrom a chemical or biochemical point of view, instrumental calibration is an essential stage in many procedures of measurement for the quantitative determination of an analyte in a sample and also to evaluate the quality of the procedure. Here Monte Carlo simulations are used to demonstrate the flaws in GOF tests. These neglect the constant variance component at small signal, leading to overly optimistic estimates of detection and quantification limits. By contrast, the simple weighting formulas selected by GOF tests often prescribe weighting as 1/x, 1/x 2, 1/y, or 1/y 2. Realistic VFs typically become constant in the low-x, low-y limit. This means that effort should be focused on estimating the data variance functions (VFs), not on these GOF tests. At the same time, a solution to this problem has been available for almost a century: The data should be weighted as their inverse variances. oldCharsize = !P.Charsize !P.charsize = 0.75 labels = line_types = REPLICATE(!P.linestyle, 5) psyms = REPLICATE(!P.psym, 5) LEGEND, labels, line_clrs, line_types, psyms, 1.6, -0.3, 0.1 !P.As analysts have become aware that their data may be strongly heteroscedastic, requiring weighted least squares in calibration fitting, many have turned to goodness-of-fit (GOF) tests as means for discerning the proper weighting formulas. !P.font = 0 TEK_COLOR clrs = WoColorConvert(INDGEN(256)) WINDOW, /Free, Xsize=700, Ysize=400 minY = MIN(, Max=maxY) PLOT, domain, P40, /Nodata, Xrange=, $ Yrange=, Ystyle=1, Xstyle=1, $ Title='4!eth!N-degree Lagrange Basis Polynomials', $ Xtitle='Abscissas: 0, 1.5, 3.6, 7, 10', $ Color=clrs(0), Background=clrs(1) OPLOT, !X.Crange,, Linestyle=1, Color=clrs(15) OPLOT, !X.Crange,, Linestyle=1, Color=clrs(15) FOR i=0L,4 DO OPLOT,, !Y.Crange, $ Linestyle=1, Color=clrs(15) line_clrs = clrs() OPLOT, domain, P40, Thick=2, Color=line_clrs(0) OPLOT, domain, P41, Thick=2, Color=line_clrs(1) OPLOT, domain, P42, Thick=2, Color=line_clrs(2) OPLOT, domain, P43, Thick=2, Color=line_clrs(3) OPLOT, domain, P44, Thick=2, Color=line_clrs(4) Add a legend. domain = INTERPOL(, 1000) P40 = POLY(domain, L40) P41 = POLY(domain, L41) P42 = POLY(domain, L42) P43 = POLY(domain, L43) P44 = POLY(domain, L44) Plot each of the basis polynomials. y = L41 = LAGRANGE(x, y) y = L42 = LAGRANGE(x, y) y = L43 = LAGRANGE(x, y) y = L44 = LAGRANGE(x, y) Now evaluate the five Lagrange basis polynomials over the domain x=. y = L40 = LAGRANGE(x, y) L40 means the 4th-degree Lagrange basis polynomial associated with the 0th abscissa. Likewise, the polynomial associated with the 1st abscissa will equal 1 at x(1)=1.5, while the other four polynomials will equal 0, etc. By definition, the polynomial associated with the 0th abscissa will equal 1 at x(0)=0, while the other four polynomials will equal 0. x = Define the function values associated with each of the five Lagrange basis polynomials.
Example 2 Make up five abscissas to define five, 4th-degree, Lagrange basis polynomials on.


 0 kommentar(er)
0 kommentar(er)
